Indole test
This article needs additional citations for verification. (November 2022) |
The indole test is a biochemical test performed on bacterial species to determine the ability of the organism to convert tryptophan into indole. This division is performed by a chain of a number of different intracellular enzymes, a system generally referred to as "tryptophanase."[citation needed]
Biochemistry
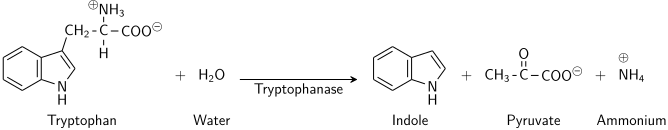
Indole is generated by reductive deamination from tryptophan via the intermediate molecule indolepyruvic acid. Tryptophanase catalyzes the deamination reaction, during which the amine (-NH2) group of the tryptophan molecule is removed. Final products of the reaction are indole, pyruvic acid, ammonium (NH4+) and energy. Pyridoxal phosphate is required as a coenzyme.
Performing a test

Like many biochemical tests on bacteria, results of an indole test are indicated by a change in color following a reaction with an added reagent.
Pure bacterial culture must be grown in sterile tryptophan or peptone broth for 24–48 hours before performing the test. Following incubation, five drops of Kovac's reagent (isoamyl alcohol, para-Dimethylaminobenzaldehyde, concentrated hydrochloric acid) are added to the culture broth.
A positive result is shown by the presence of a red or reddish-violet color in the surface alcohol layer of the broth. A negative result appears yellow. A variable result can also occur, showing an orange color as a result. This is due to the presence of skatole, also known as methyl indole or methylated indole, another possible product of tryptophan degradation.
The positive red color forms as a result of a series of reactions. The para-Dimethylaminobenzaldehyde reacts with indole present in the medium to form a red rosindole dye. The isoamyl alcohol forms a complex with rosindole dye, which causes it to precipitate. The remaining alcohol and the precipitate then rise to the surface of the medium.
A variation on this test using Ehrlich's reagent (using ethyl alcohol in place of isoamyl alcohol, developed by Paul Ehrlich) is used when performing the test on nonfermenters and anaerobes.
Indole-Positive Bacteria
Bacteria that test positive for cleaving indole from tryptophan include: Aeromonas hydrophila, Aeromonas punctata, Bacillus alvei, Edwardsiella sp., Escherichia coli, Flavobacterium sp., Haemophilus influenzae, Klebsiella oxytoca, Proteus sp. (not P. mirabilis and P. penneri), Plesiomonas shigelloides, Pasteurella multocida, Pasteurella pneumotropica, Vibrio sp., and Lactobacillus reuteri.
Indole-Negative Bacteria
Bacteria which give negative results for the indole test include: Actinobacillus spp., Aeromonas salmonicida, Alcaligenes sp., most Bacillus sp., Bordetella sp., Enterobacter sp., most Haemophilus sp., most Klebsiella sp., Neisseria sp., Mannheimia haemolytica, Pasteurella ureae, Proteus mirabilis, P. penneri, Pseudomonas sp., Salmonella sp., Serratia sp., Yersinia sp., and Rhizobium sp.
The Indole test is one of the four tests of the IMViC series, which tests for evidence of an enteric bacterium. The other three tests include: the methyl red test [M], the Voges–Proskauer test [V] and the citrate test [C].[1]
References
- ^ Bachoon, Dave S., and Wendy A. Dustman. Microbiology Laboratory Manual. Ed. Michael Stranz. Mason, OH: Cengage Learning, 2008. Exercise 15, "Normal Flora of the Intestinal Tract" Print.
- MacFaddin, Jean F. "Biochemical Tests for Identification of Medical Bacteria." Williams & Wilkins, 1980, pp 173 – 183.
- Example of typical indole reactions
- Angen, O.; Mutters, R.; Caugant, D. A.; Olsen, J. E.; Bisgaard, M. (1999). "Taxonomic relationships of the [Pasteurella] haemolytica complex as evaluated by DNA-DNA hybridizations and 16S rRNA sequencing with proposal of Mannheimia haemolytica gen. nov., comb, nov., Mannheimia granulomatis comb. nov., Mannheimia glucosida sp. nov., Mannheimia ruminalis sp. nov. and Mannheimia varigena sp. nov.". International Journal of Systematic Bacteriology. 49 (1): 67–86. doi:10.1099/00207713-49-1-67. ISSN 0020-7713. PMID 10028248
